Help
| 1. Dataset of ChimerDB 3.0 |
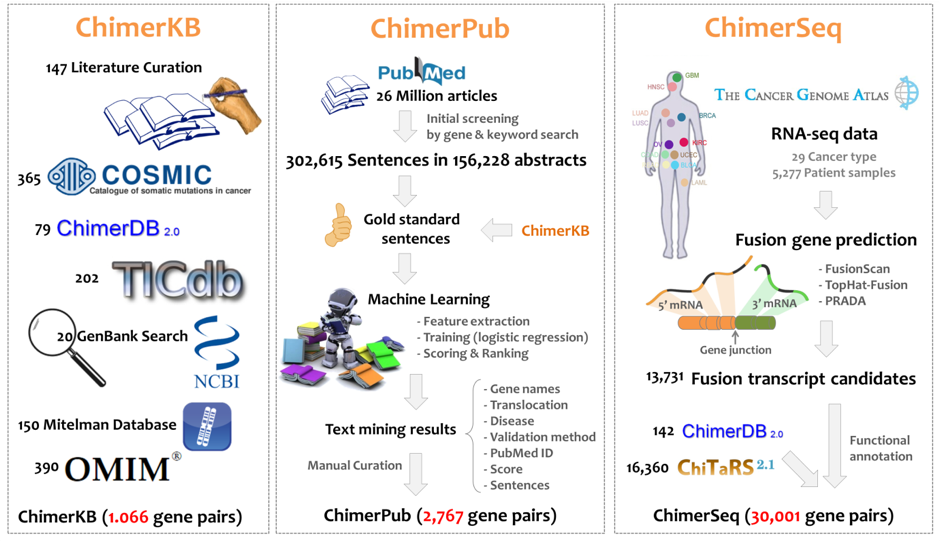
|
| 1.1 ChimerKB |
| ChimerKB represents a knowledgebase including 1,066 fusion genes that were manually curated and compiled from public resources of fusion genes based on experimental results. Fusion genes were compiled from well-known public resources such as GenBank, Mitelman, OMIM, COSMIC, TICdb and PubMed articles. All entries were manually curated for disease, sequences, break points, and experimental evidences. |
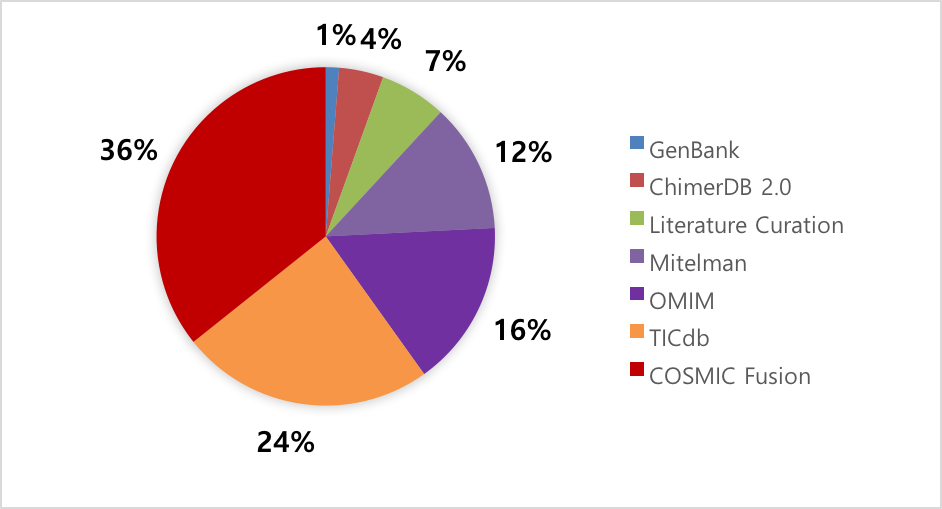
|
| Kim, P., Yoon, S., Kim, N., Lee, S., Ko, M., Lee, H., ... & Lee, S. (2010). ChimerDB 2.0—a knowledgebase for fusion genes updated. Nucleic acids research, 38(suppl 1), D81-D85. | Benson, D. A., Cavanaugh, M., Clark, K., Karsch-Mizrachi, I., Lipman, D. J., Ostell, J., & Sayers, E. W. (2013). GenBank. Nucleic acids research, 41(D1), D36-D42. http://www.ncbi.nlm.nih.gov/genbank/ |
| Mitelman, F., Johansson, B., & Mertens, F. (2007). Mitelman database of chromosome aberrations in cancer. Cancer Genome Anatomy Project. http://cgap.nci.nih.gov/Chromosomes/Mitelman |
| Hamosh, A., Scott, A. F., Amberger, J. S., Bocchini, C. A., & McKusick, V. A. (2005). Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic acids research, 33(suppl 1), D514-D517. http://www.omim.org/ |
| Novo, F. J., de Mendíbil, I. O., & Vizmanos, J. L. (2007). TICdb: a collection of gene-mapped translocation breakpoints in cancer. BMC genomics, 8(1), 1. http://www.unav.es/genetica/TICdb/ |
| 1.2 ChimerPub |
|
ChimerPub provides up-to-date information on published fusion genes.
We developed an advanced text mining system to identify fusion genes.
To obtain fusion gene information from PubMed articles, we first searched for all the sentences
that containing at least two co-occurring genes using the BEST entity extractor [BEST Citation].
We collected a total of 302,615 sentences in 156,228 abstracts published before June 2016.
Through manual curation, we constructed a gold-standard fusion gene sentence set of 1,549 sentences.
We extracted features from the sentences including differentially distributed words for machine
learning and score and ranks of all target sentences with logistic regression.
Among the top 10,000 scored sentences, 2,563 unique fusion genes were identified, and 2,067 of them which were novel.
|
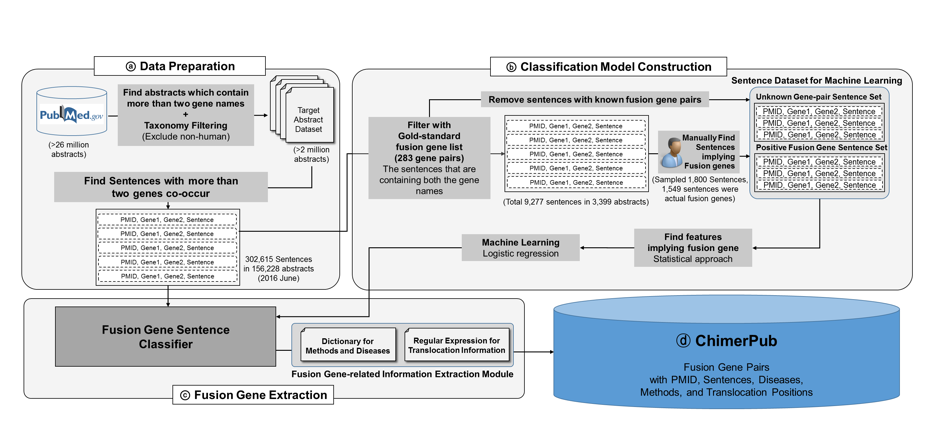
|
|
Fig. 1 Overall workflow of ChimerPub
ⓐ From all the PubMed articles, 302,615 sentences in 156,228 abstracts were chosen as fusion gene candidate sentences.
ⓑ Using a gold-standard fusion gene list and manual curation, we collected 1,549 sentences for the positive fusion gene set,
which were used for machine learning.
ⓒ The trained machine learning model was used for identify fusion genes from all the candidate sentences.
Related information included extraction module extracts diseases, experimental methods, and translocation positions.
This process is automated so that ChimerPub can provide updated fusion gene information every day.
ⓓ The organized fusion gene pairs and their related information are provided in ChimerPub.
|
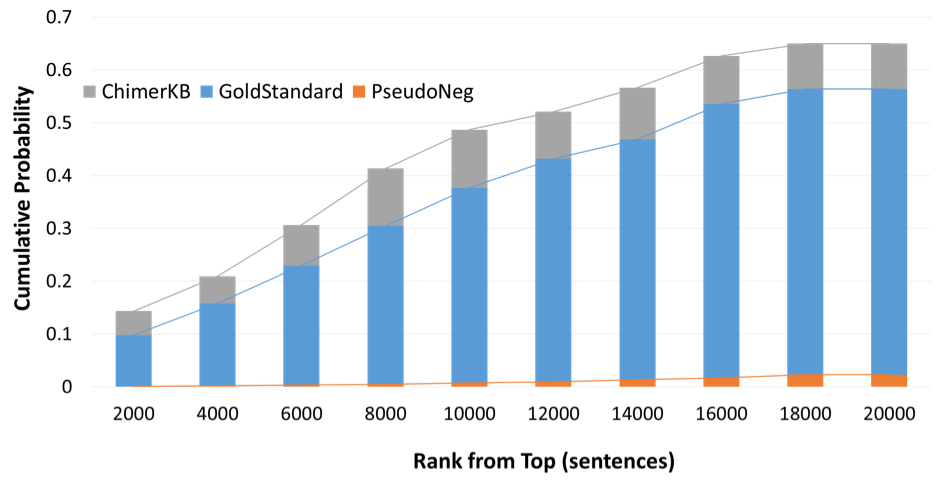
|
|
Fig. 2 Reliability of ChimerPub text-mining result
The graph indicates the cumulative probability of including entries of each resources.
GoldStandard and PseudoNeg are the sets of sentences used in the training process.
ChimerKB entries are pseudo-positive sentences containing gene names of genuine the fusion genes from ChimerKB.
For example, top 10,000 sentences include almost 50% of ChimerKB-generated sentences.
|
| 1.3 ChimerSeq |
|
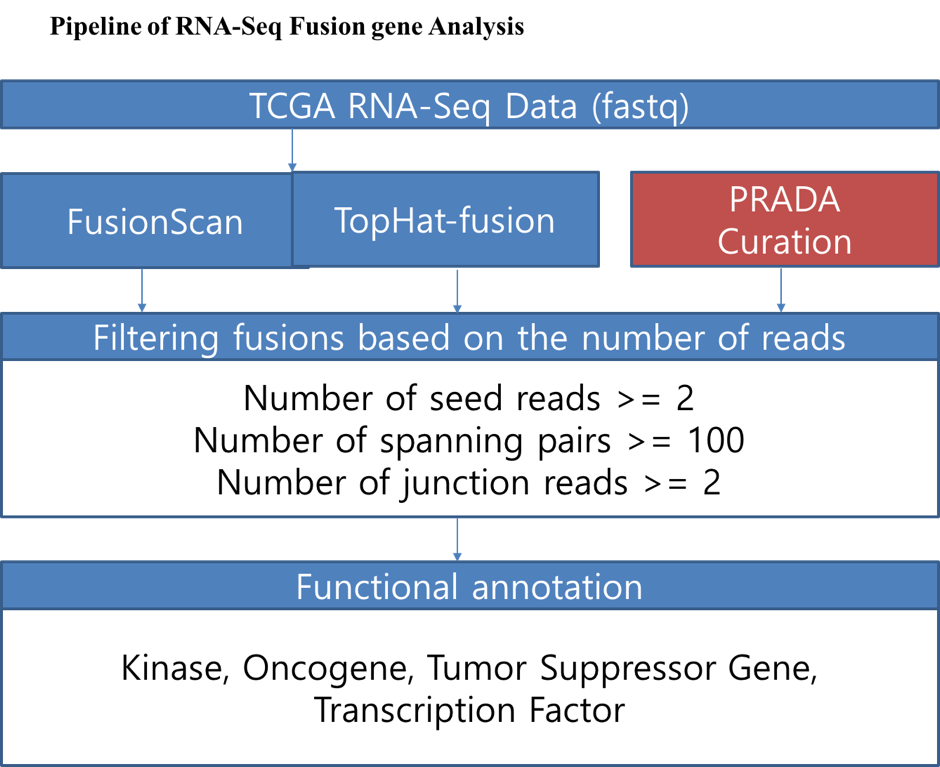
Raw RNA-seq data were downloaded from the TCGA data portal with the permission of dbGAP.
Because TopHat-Fusion and FusionScan archieved the best performance in our fusion tools performance test,
we analyzed the TCGA RNA-seq data using FusionScan and TopHat-Fusion.
The Fusion gene list of TCGA samples from the PRADA pipeline was also included in our database(http://www.tumorfusions.org).
Fusion transcripts detected by analyzing Human ESTs and mRNAs were curated from ChiTaRS-2.1 and
Sequence Read Archive (SRA) predictions of confidence Class A from ChimerDB 2.0.
|
| Kim, D., & Salzberg, S. L. (2011). TopHat-Fusion: an algorithm for discovery of novel fusion transcripts. Genome Biol, 12(8), R72. | Yoshihara, K., Wang, Q., Torres-Garcia, W., Zheng, S., Vegesna, R., Kim, H., & Verhaak, R. G. W. (2014). The landscape and therapeutic relevance of cancer-associated transcript fusions. Oncogene. http://www.tumorfusions.org. |
| Frenkel-Morgenstern, M., Gorohovski, A., Lacroix, V., Rogers, M., Ibanez, K., Boullosa, C., ... & Valencia, A. (2013). ChiTaRS: a database of human, mouse and fruit fly chimeric transcripts and RNA-sequencing data. Nucleic acids research, 41(D1), D142-D151. http://chitars.bioinfo.cnio.es/ |
| 1.3.1 Data processings |
| TopHat-Fusion |
| TCGA RNA-seq data reads were aligned to the genome reference UCSC hg19 refGene and ensGene with Bowtie1. | tophat -p 8 --fusion-search --keep-fasta-order --bowtie1 --no-coverage-search -r 0 --mate-std-dev 80 --max-intron-length 100000 --fusion-min-dist 100000 --fusion-anchor-length 13 --fusion-ignore-chromosomes chrM |
| FusionScan |
| SSAHA2 was used to map TCGA RNA-Seq reads to the human transcriptome of refGene from the UCSC genome annotation database for the hg19 (GRCh37). |
| python Run_FusionScan.py TCGA-PK-A5HB-01A_1.fastq,TCGA-PK-A5HB-01A_2.fastq TCGA-PK-A5HB-01A 48 10.10.20.106:8652 --phred33 -P 8 -ms 2 |
 |
| 1.3.2 FusionScan |
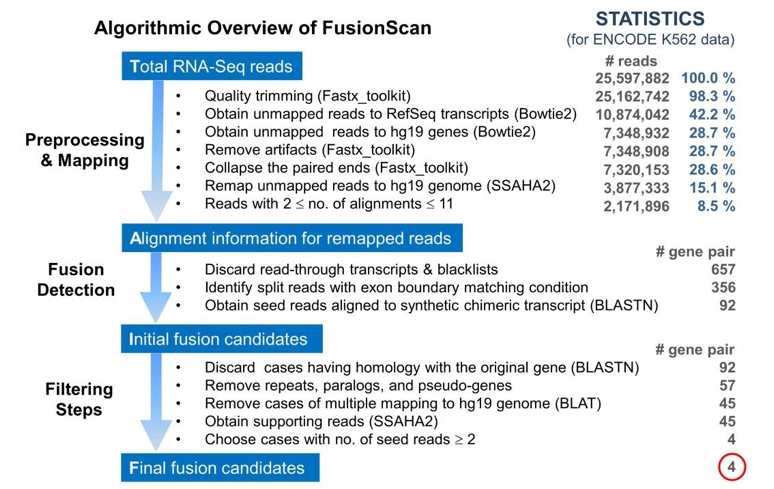
| FusionScan, a highly optimized tool for predicting fusion transcripts from RNA-Seq data. We specifically searched for split reads composed of intact exons at the fusion boundaries. We implemented various mapping and filtering strategies to remove false-positives without discarding genuine cases. FusionScan outperformed existing programs by a considerable margin, achieving precision and recall rates of 60% and 79%, respectively. |
 |
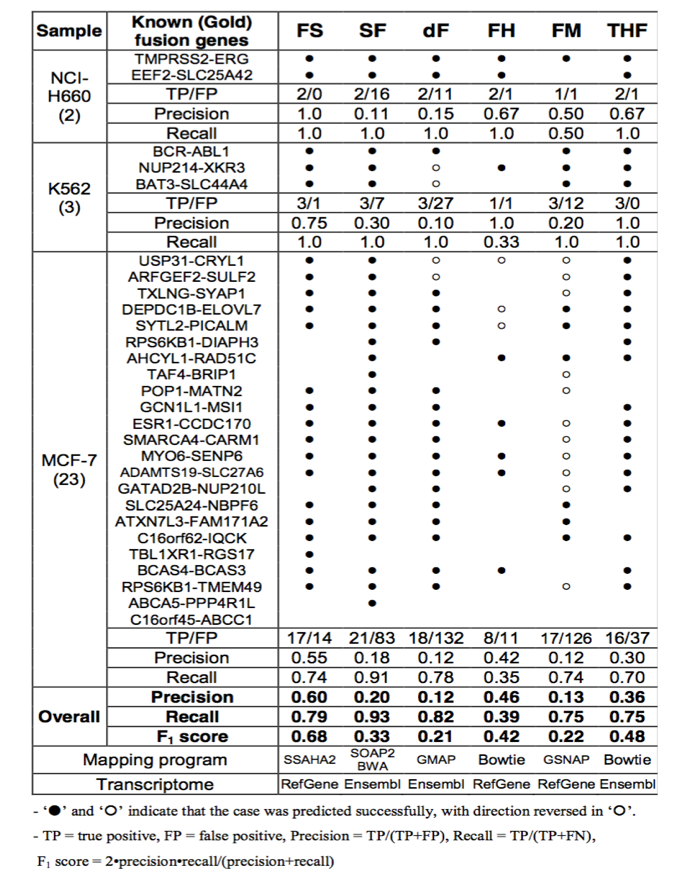
| Summary of known fusion genes detected by each tool and the comparison statistics. |
 |
| FusionScan Documentation |
| Go to FusionScan Web page |
| 2. ChimerKB Menu |
| 2.1 How-to search |
|
Please select a feature (search type: Gene, Gene-pair, Disease, show all entries) first. When selected,
|
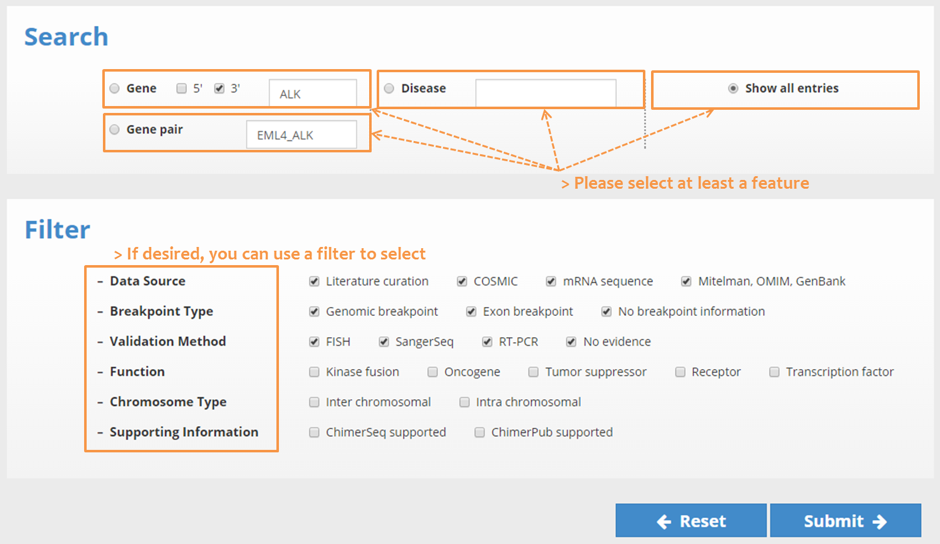 |
| Select the filter conditions. (Data Source, Breakpoint Type, Validation Method, Function, Chromosome Type, Supporting Information) Specify the criteria to use to filter the displayed data. |
| 2.2 Search result: Brief information & detailed information |
| Output GUI consists of a table with search hits, detailed information regarding a specific fusion event. The output table supports many features of searching, sorting, exporting, and link outs to external resources. There were ten data fields as fusion gene, 5’gene junction, 3’gene junction, breakpoint type, disease, frame, chromosome information, data source, supporting Information, PubMed ID. Click a row to show the detailed information related to the fusion gene of interest. The table below describes the selected fusion gene. The PubMed Id links are connected; go to the ChimerPub, which provide supporting information regarding the link connections. |
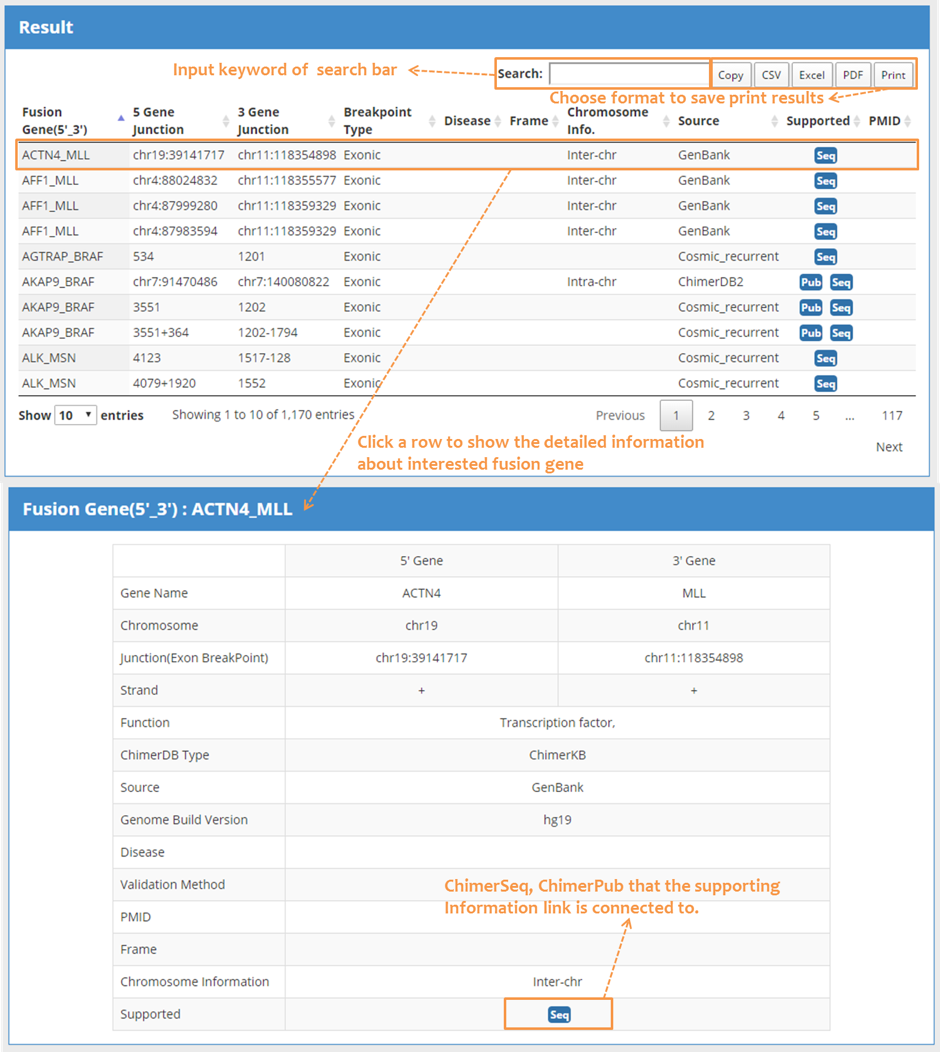 |
| 3. ChimerPub Menu |
| 3.1 How-to search |
| The ChimerPub search is connected in the same manner as described above. The number of publications and text mining score were added to the filters. |
 |
| 3.2 Search result: Brief information and abstract highlights |
| There were six data fields: fusion gene, translocation, function, disease, supporting Information, PubMed ID. Click a row to show the abstract information for the fusion gene of interest. The table below describes the extracted fusion gene obtained by text mining of the literature. |
 |
| 4. ChimerSeq Menu |
| 4.1 How-to search |
| We support diverse types of searching, including gene, gene pair, chromosome locus, and disease types. In ChimerSeq search, users may select the data source, cancer type, and prediction tools with optional parameters. With ample annotations, we support diverse filtering options such as function filter for kinase, oncogene, tumor suppressor, receptor, transcription factor genes. |
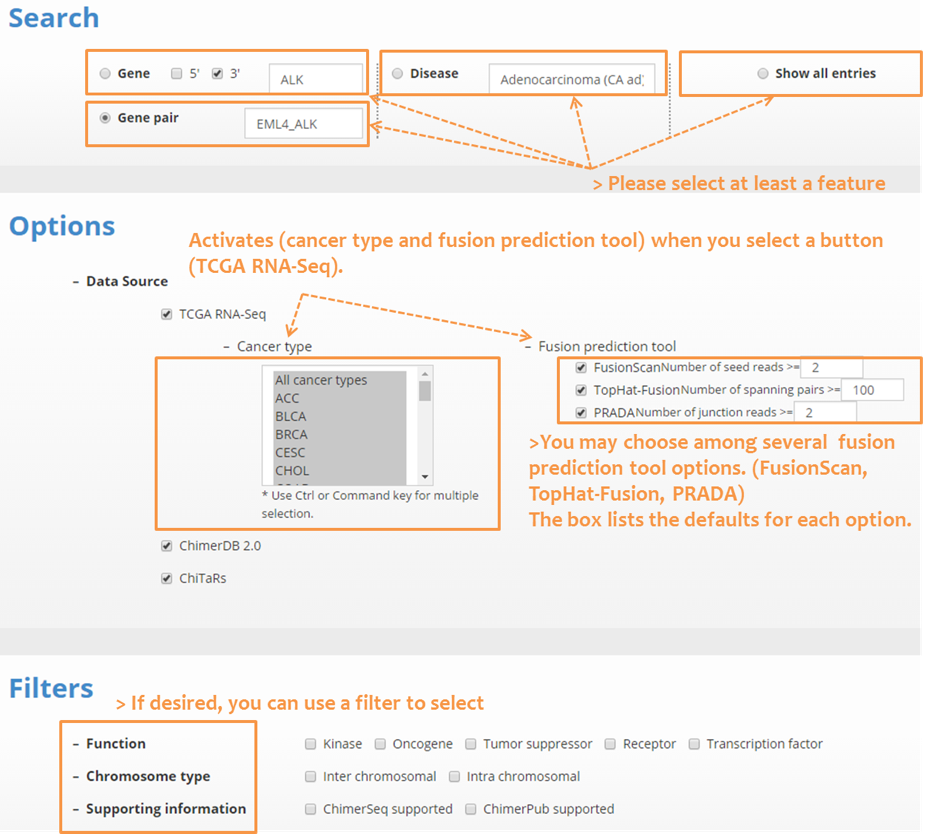 |
| 4.2 Search result: Brief information & detailed information |
| The output GUI consists of a summary table with search hits, a graphic illustration of fusion structure, and detailed information regarding a specific fusion event. The output table supports many features of searching, sorting, exporting, and link outs to external resources. There were nine data fields: fusion gene, 5’gene junction, 3’gene junction, breakpoint type, cancer type, frame, chromosome information, data source, supporting Information. Click on each entry activates the detailed information table. |
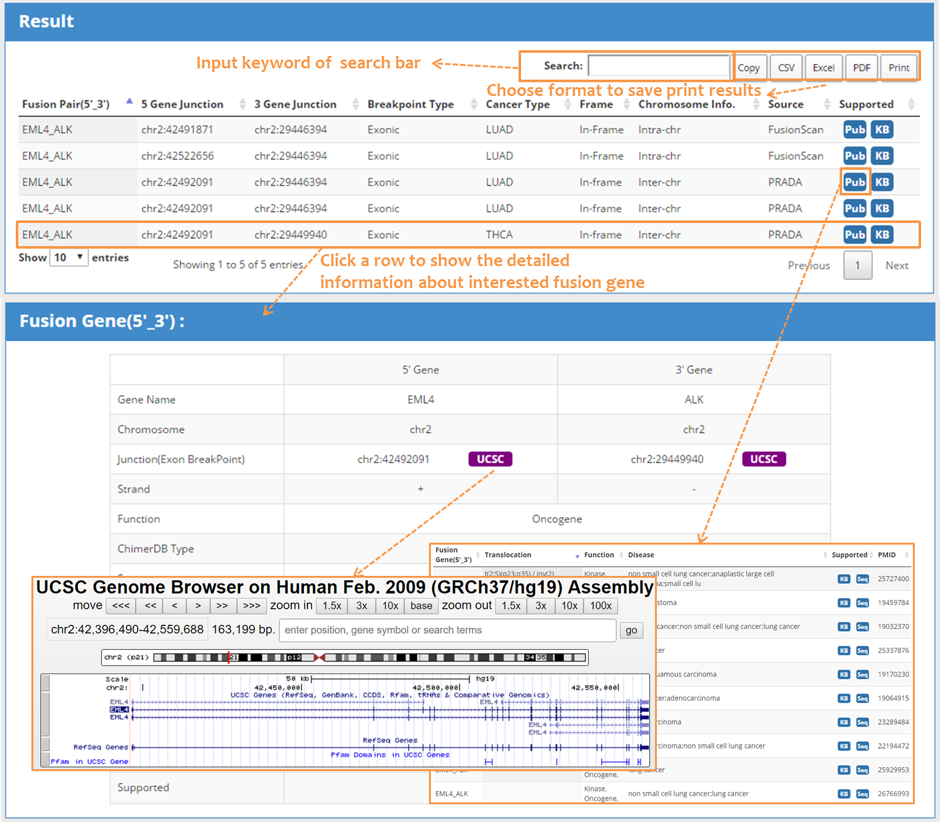 |
| 4.3 Search result: Fusion Structure |
| Click on each entry activates the graphic window of fusion gene structure. The fusion gene graphic window shows the exons, domains, and the break point before and after the fusion event. This is the most informative picture for deducing the functional significance of fusion event. If available, the alignment of short reads (seed/junction read only) is shown. Zooming and panning are supported for user convenience. The detailed information table provides all relevant information related to the fusion transcript. Notably, the UCSC links guide users to the UCSC genome browser with short read alignment as a custom track so that they can examine the detailed gene structure and alignment. |
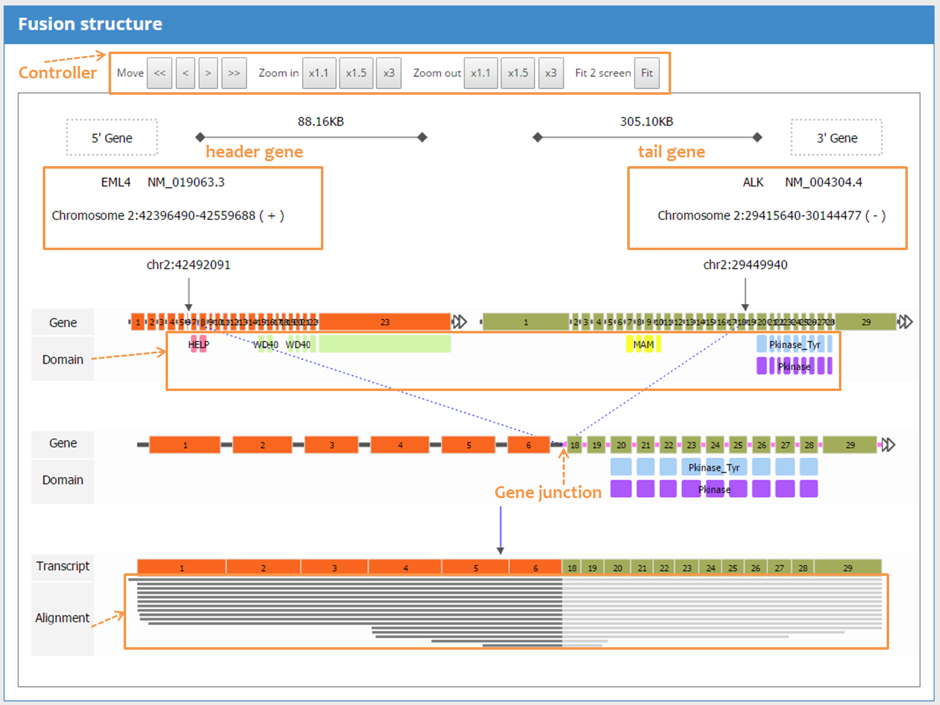 |