About ChimerDB
ChimerDB is a comprehensive database of fusion genes encompassing analysis of deep sequencing data and manual curations. In this update, the database coverage was enhanced considerably by adding two new modules of TCGA RNA-Seq analysis and PubMed abstract mining.
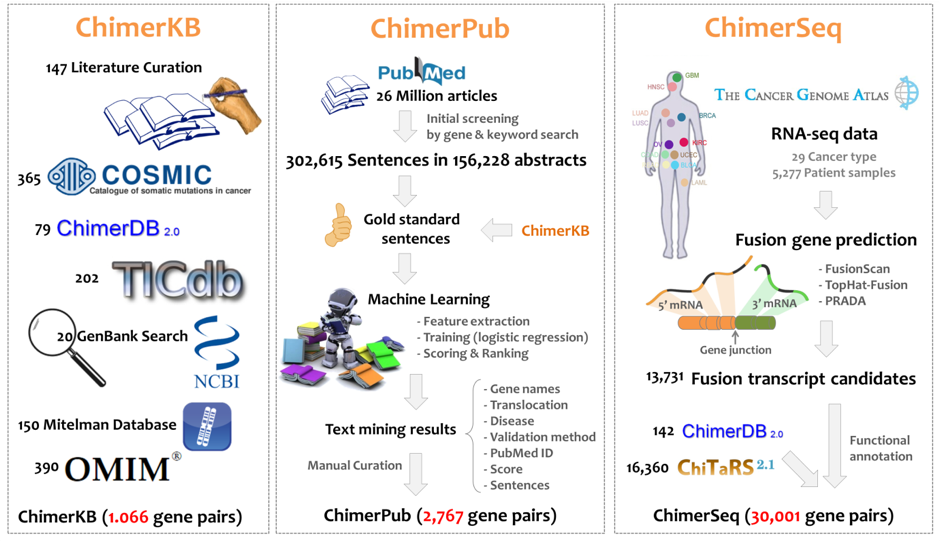
Major Features
ChimerDB 3.0 is composed of three modules of ChimerKB, ChimerPub, and ChimerSeq.
- ChimerKB represents a knowledgebase including 1,066 fusion genes with manual curation that were compiled from public resources of fusion genes with experimental evidences.
- ChimerPub includes 2,767 fusion genes obtained from text mining of PubMed abstracts.
- ChimerSeq module is designed to archive the fusion candidates from deep sequencing data. Importantly, we have analyzed RNA-Seq data of the TCGA project covering 4,569 patients in 23 cancer types using two reliable programs of FusionScan and TopHat-Fusion.
References
- ChimerDB 3.0 : an enhanced database of fusion genes with cancer transcriptome and literature data mining. in press
- ChimerDB 2.0 : a knowledgebase for fusion genes updated. Kim P, Yoon S, Kim N, Lee S, Ko M, Lee H, Kang H, Kim J, Lee S. Nucleic Acids Res. 2010 Jan;38(Database issue):D81-5. Website
- ChimerDB : a knowledgebase for fusion sequences. Kim N, Kim P, Nam S, Shin S, Lee S. Nucleic Acids Res. 2006 Jan 1;34(Database issue):D21-4. Website