Help
| 1. Overall distribution of ChimerDB 3.0 fusion genes |
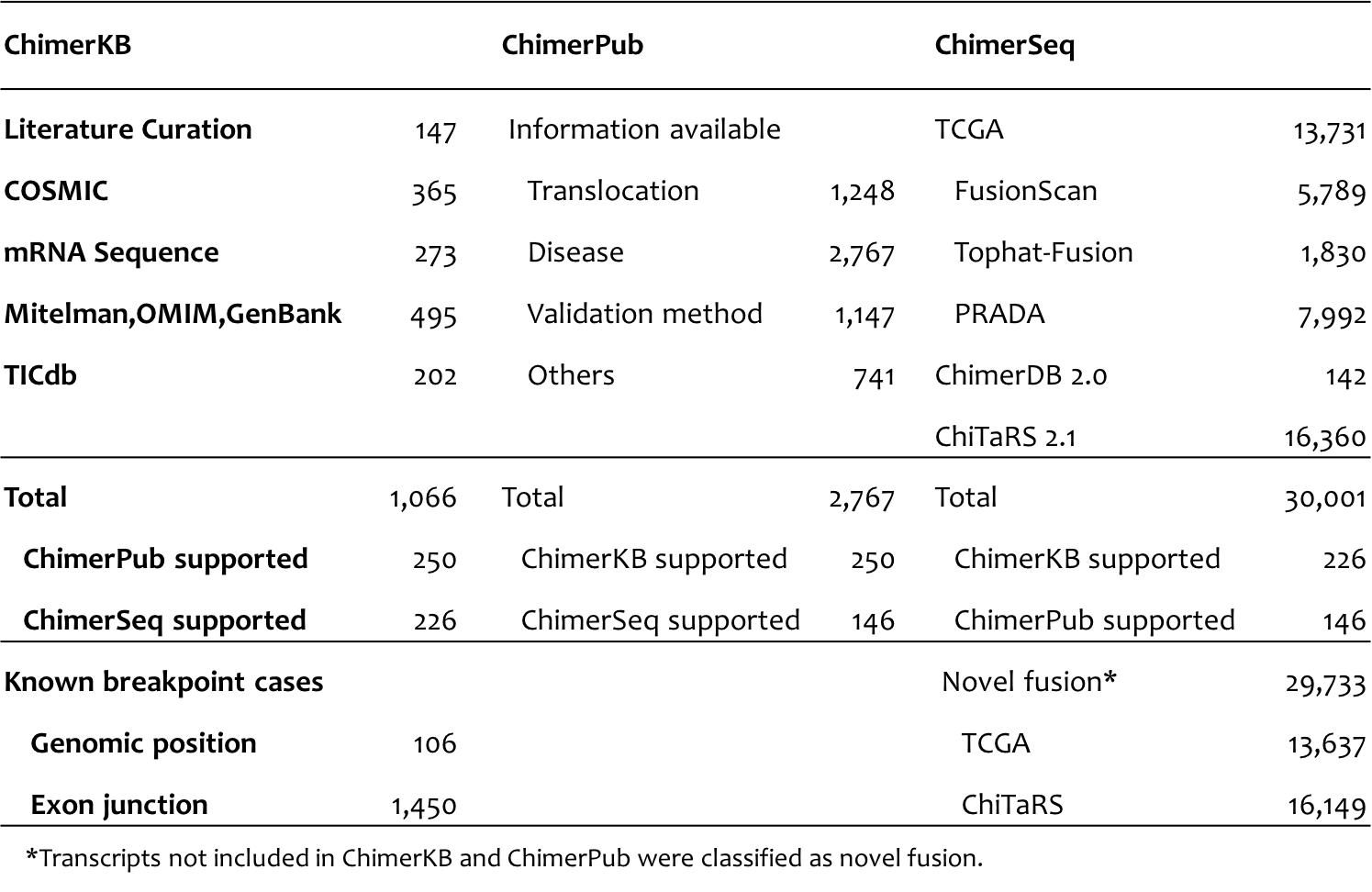
|
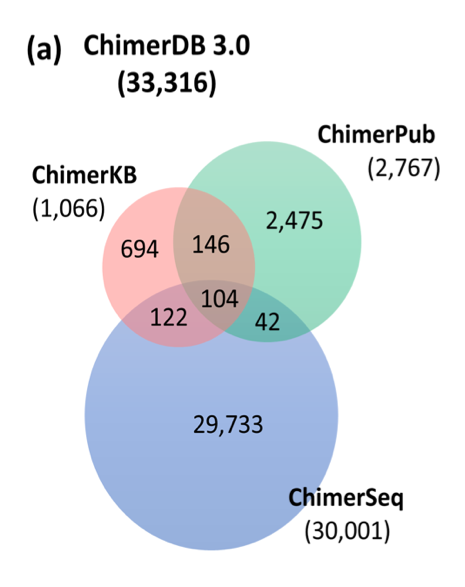
|
| ChimerDB 3.0 is composed of three main data sources of fusion genes which are ChimerKB, ChimerSeq, ChimerPub. ChimerKB include manually curated and 1,066 fusion genes with experimental evidences from COSMIC, GenBank, OMIM, Mitelman, TICdb, and ChimerDB 2.0. |
| ChimerSeq is a collection of 30,001 fusion transcripts that we have analyzed the TCGA transcriptome sequencing data using fusion gene predicting programs FusionScan and TopHat-Fusion. About 8000 PRADA analysis results and ~16000 curated fusion cases from ChiTaRS were also included in ChimerSeq database. |
| Venn diagram (a) show the detail fusion gene distribution of ChimerDB 3.0 according to each source ChimerKB, ChimerPub, ChimerSeq. As shown as the first Venn diagram, there were 33,316 unique fusion gene pairs and 104 common gene pairs among all sources in ChimerDB 3.o. |
| 2. Overall distribution of ChimerDB 3.0 fusion genes |
|
| Venn diagram(b) describes that comparison of overlapped fusion genes between TCGA fusion gene analysis algorithm. Top-hat-fusion has the smallest number of total fusion genes among three programs. |
| 3. Reliability of ChimerPub text-mining result |
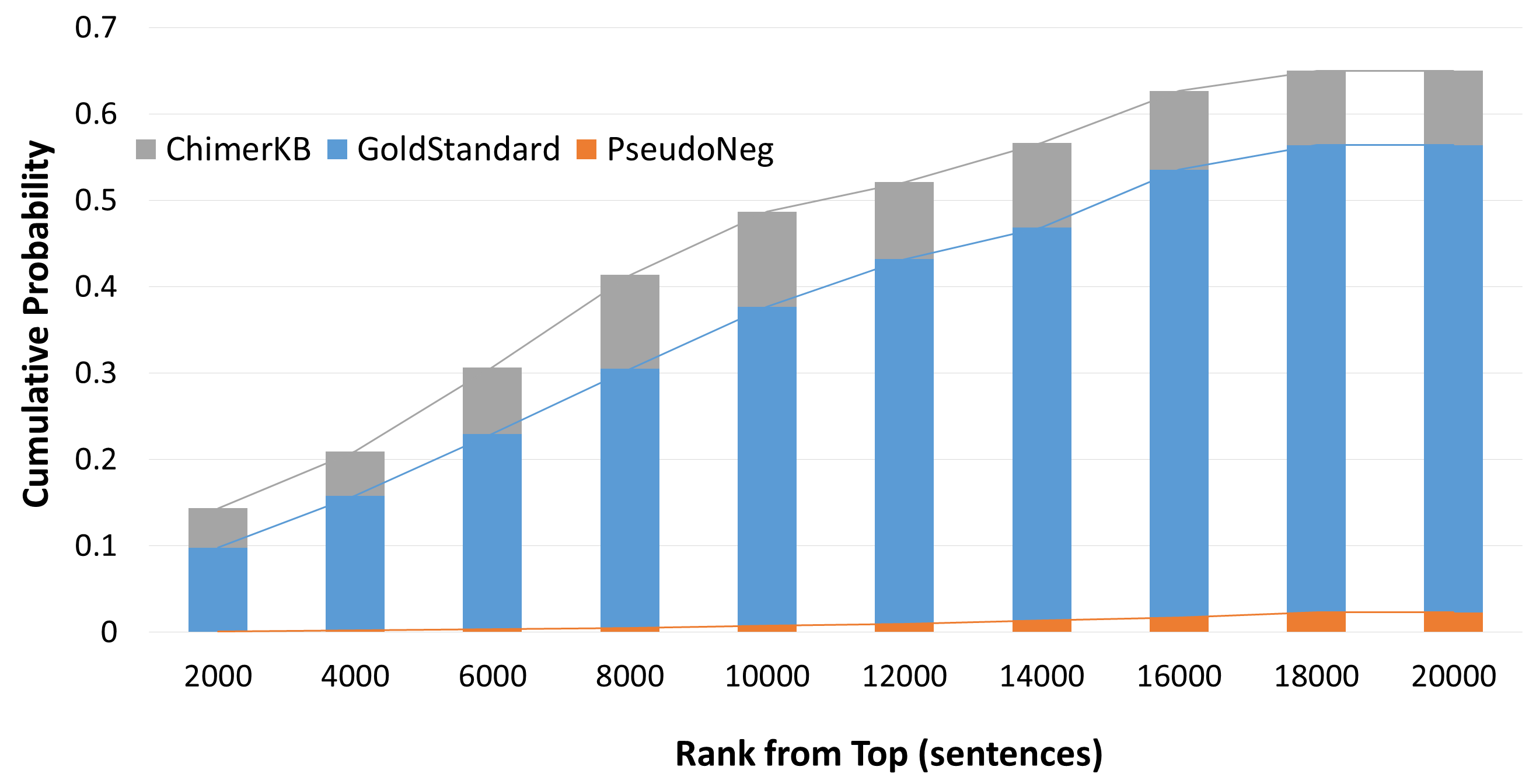
|
| The graph indicates the cumulative probability of including entries of each resources. GoldStandard and PsudoNeg are the sets of sentences used in the training process. ChimerKB entries are pseudo-positive sentences containing gene names of genuine the fusion genes from CHimerKB. |
| 4. Distribution of fusion transcripts for each cancer type |
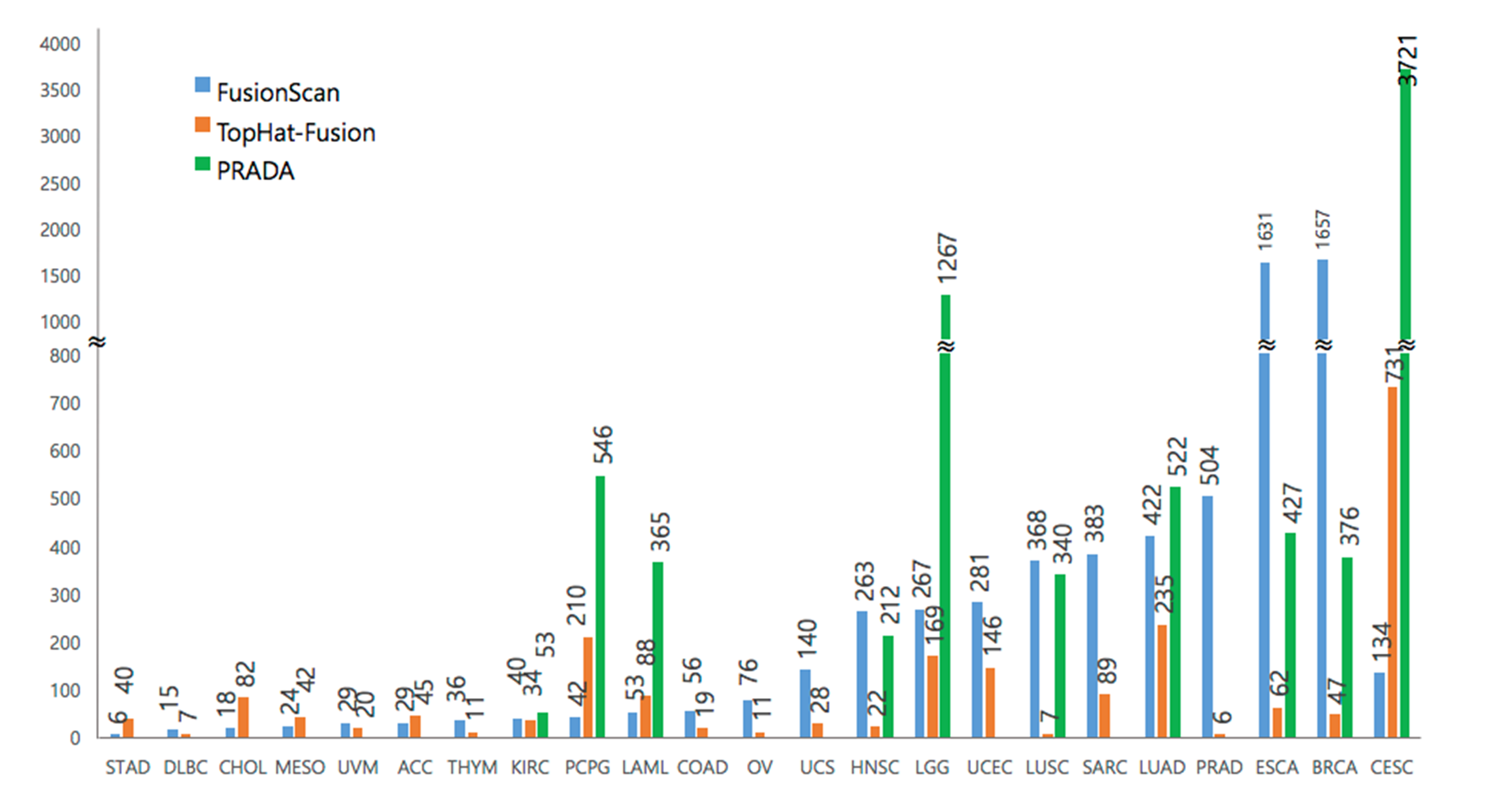
|
| This bar chart depicts the number of fusion transcripts from each algorithm uses in ChimerSeq. The blue one indicates Fusion Scan results, Orange bar is the results of Top-Hat-Fusion and green bar shows the act of fusion cases with PRADA algorithm. The largest number of fusion transcripts were produced by PRADA from Cervical cancer. The average number of transcripts is 293. |