- 1. Search
- 1-1. Screen Selector
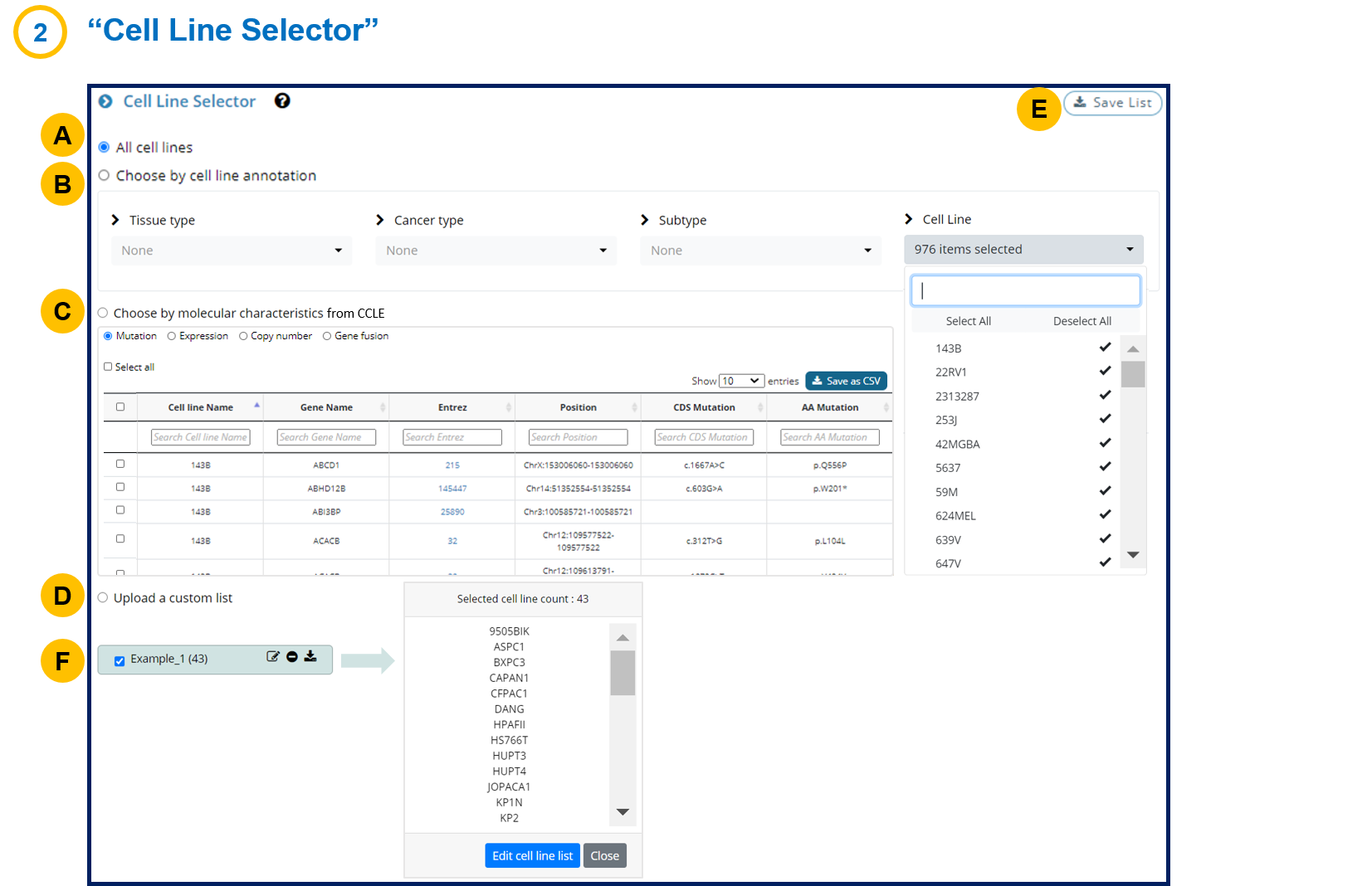
- 1-2. Cell Line Selector
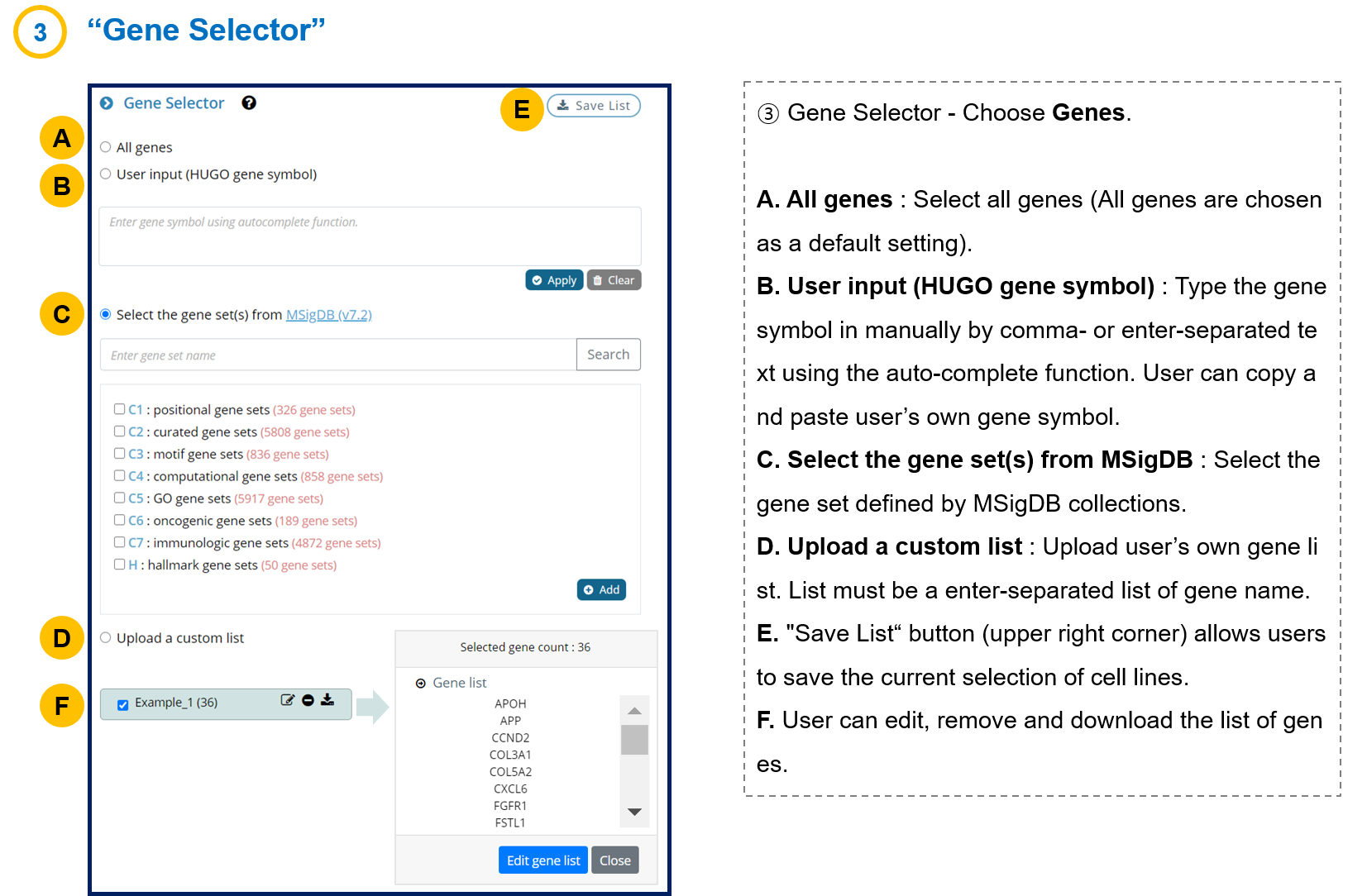
- 1-3. Gene Selector
- 1-4. Example
- 1-5. Search CRISPR screen
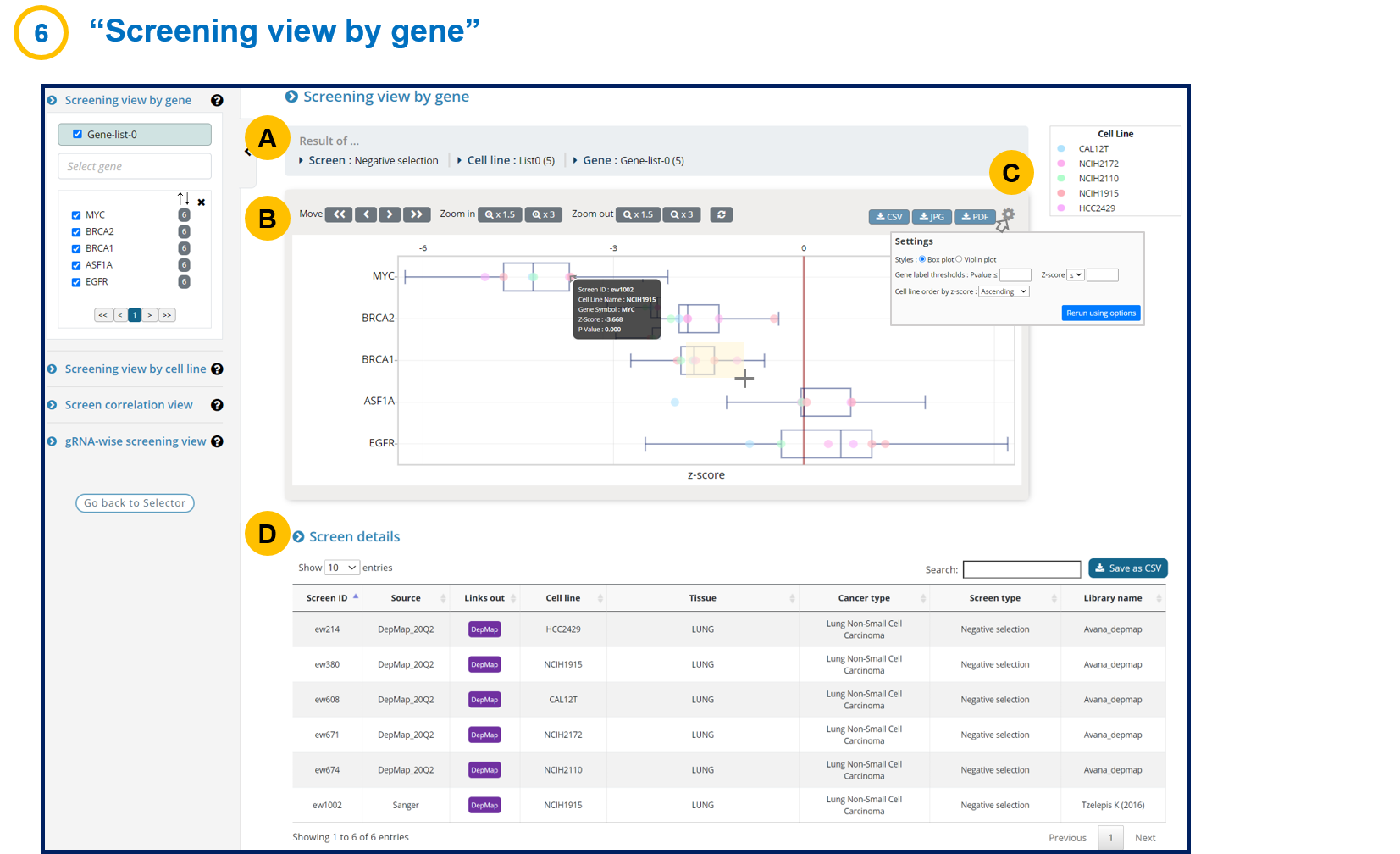
- 1-6. Screening view by gene
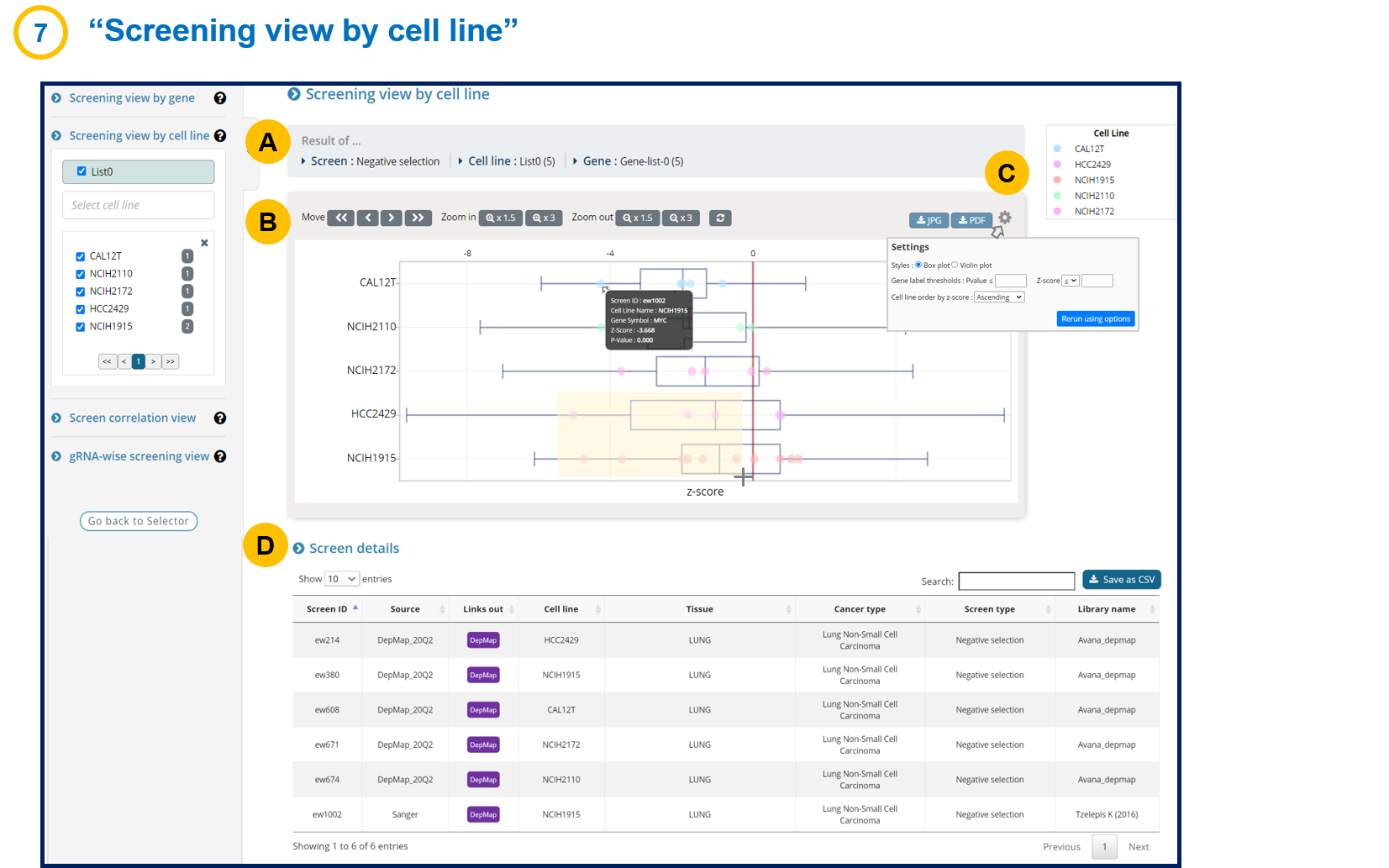
- 1-7. Screening view by cell line
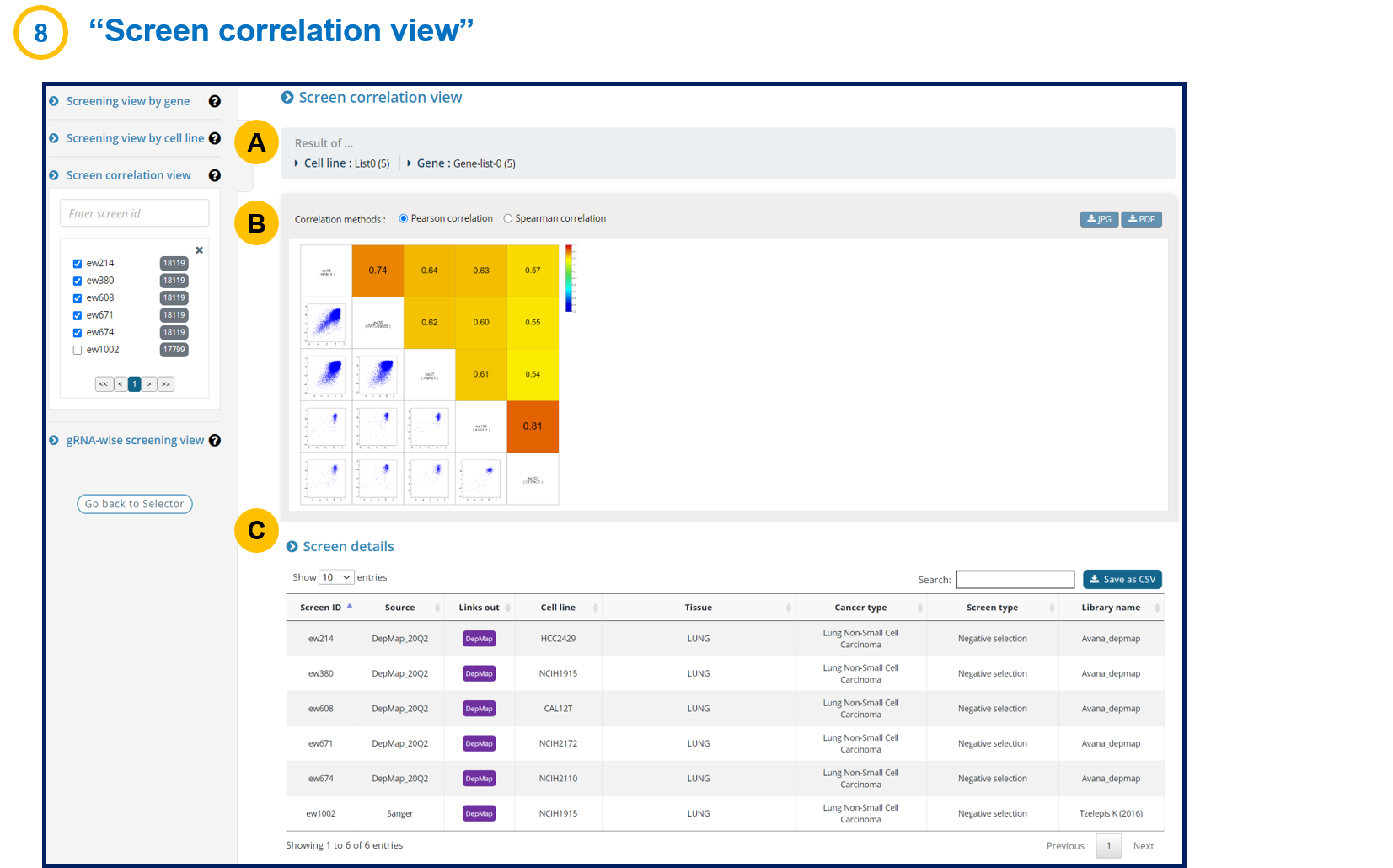
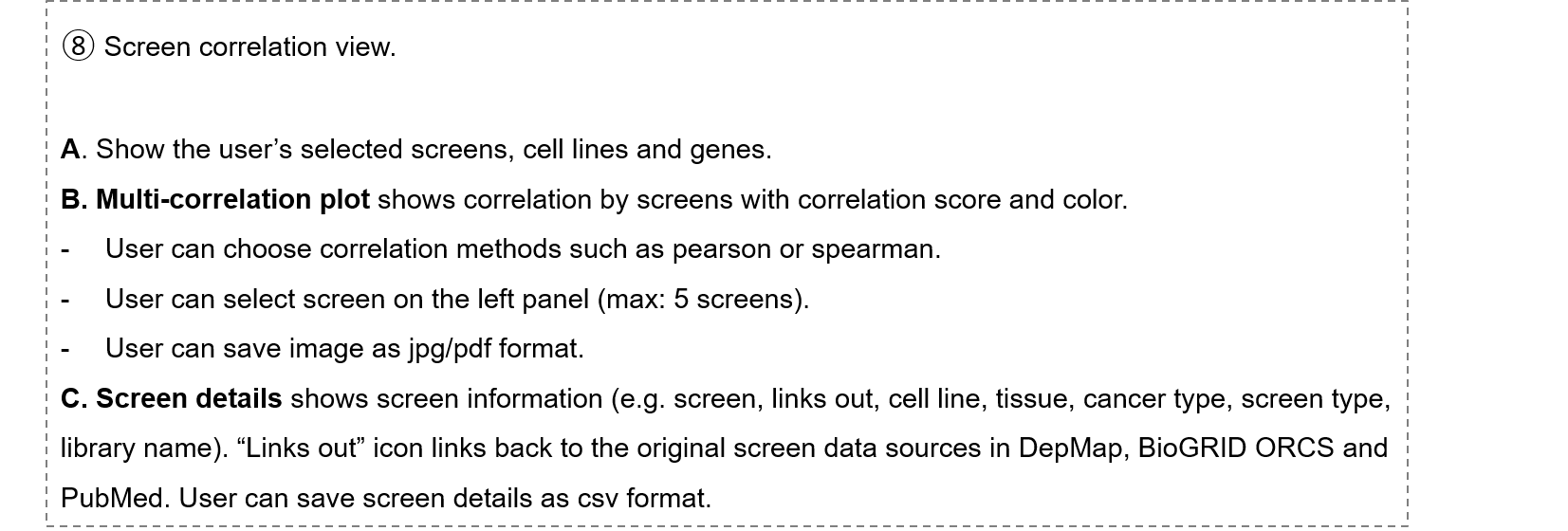
- 1-8. Screen correlation view
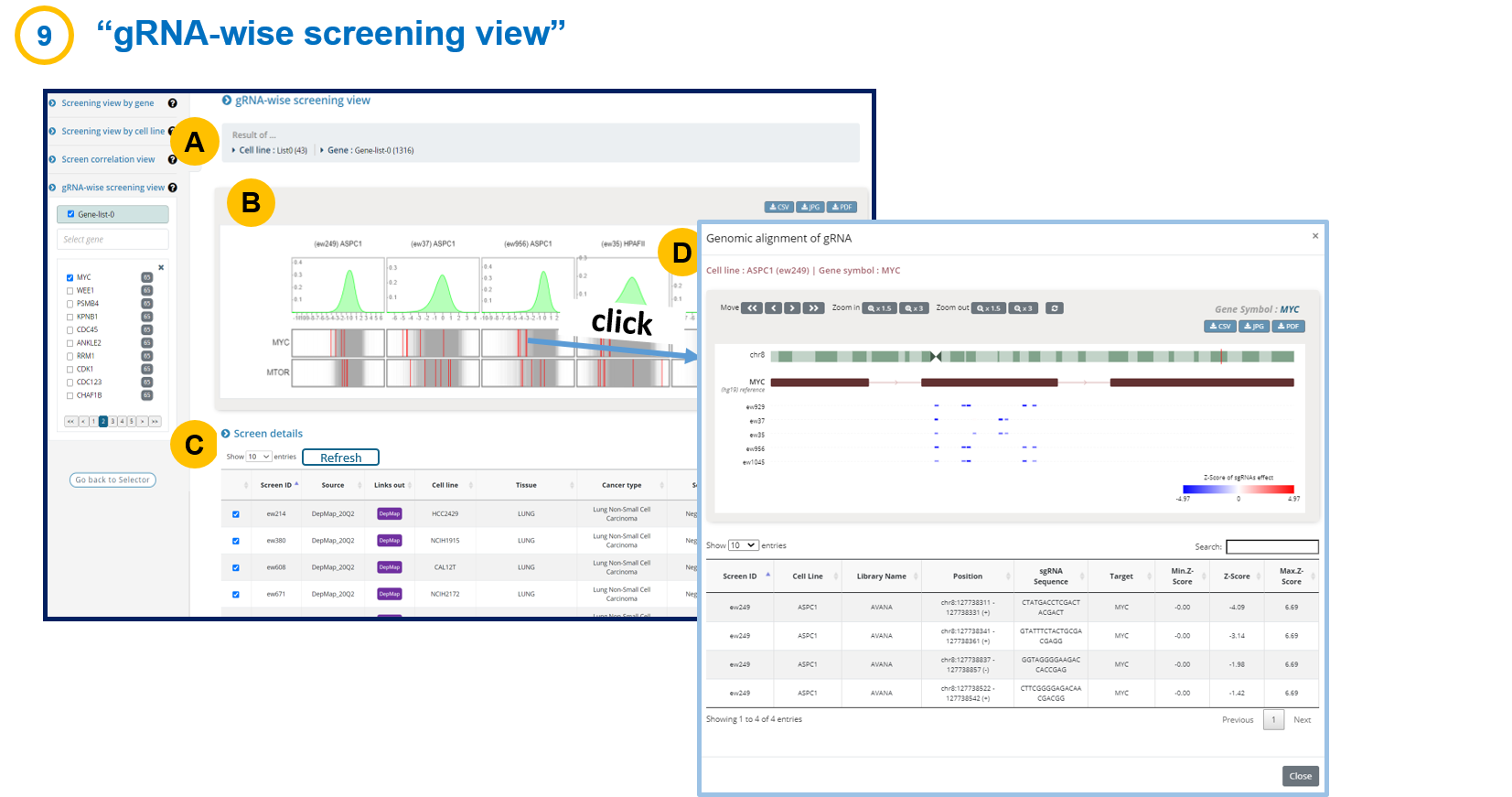
- 1-9. gRNA-wise screening view
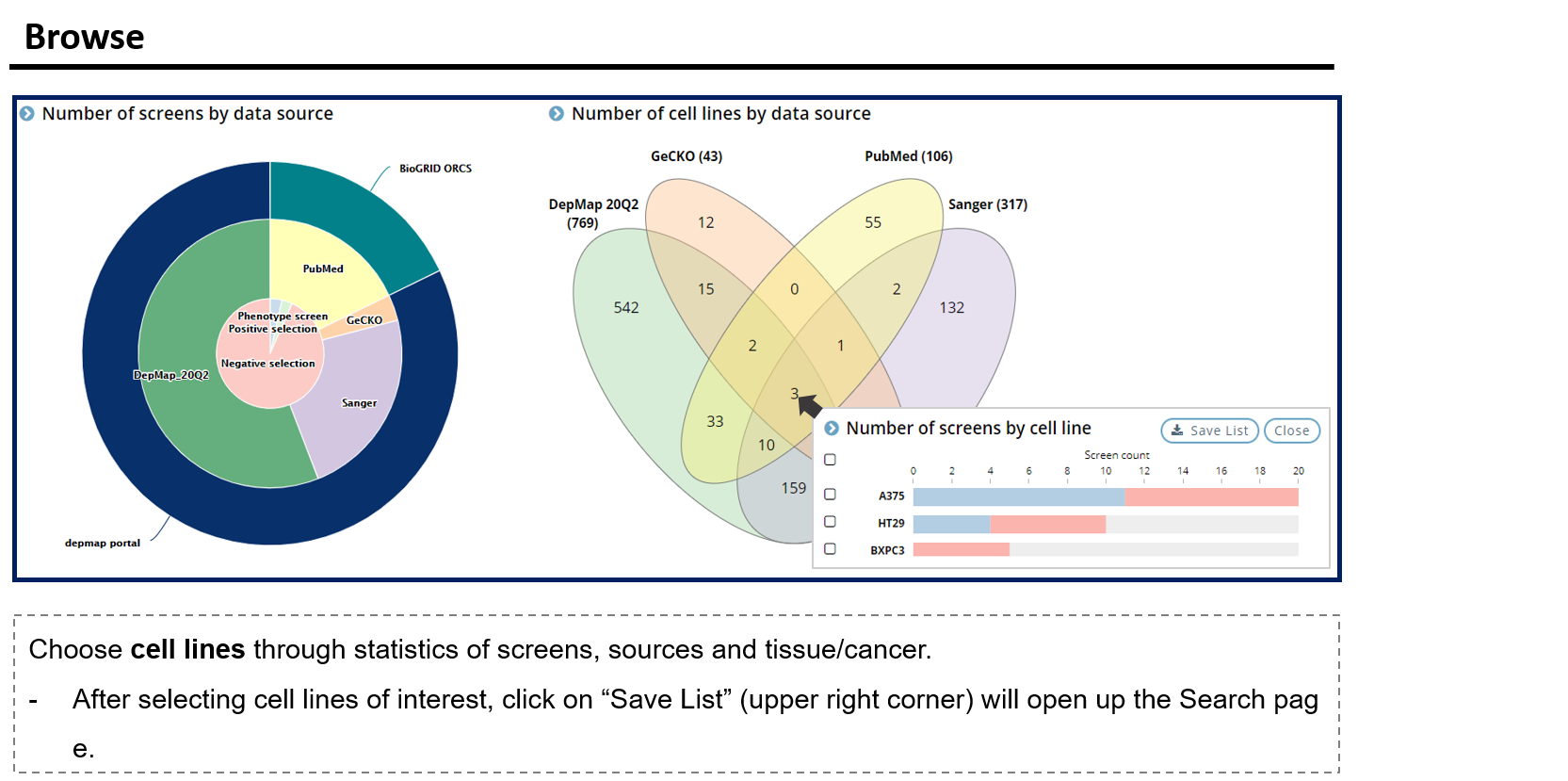
- 2. Browse
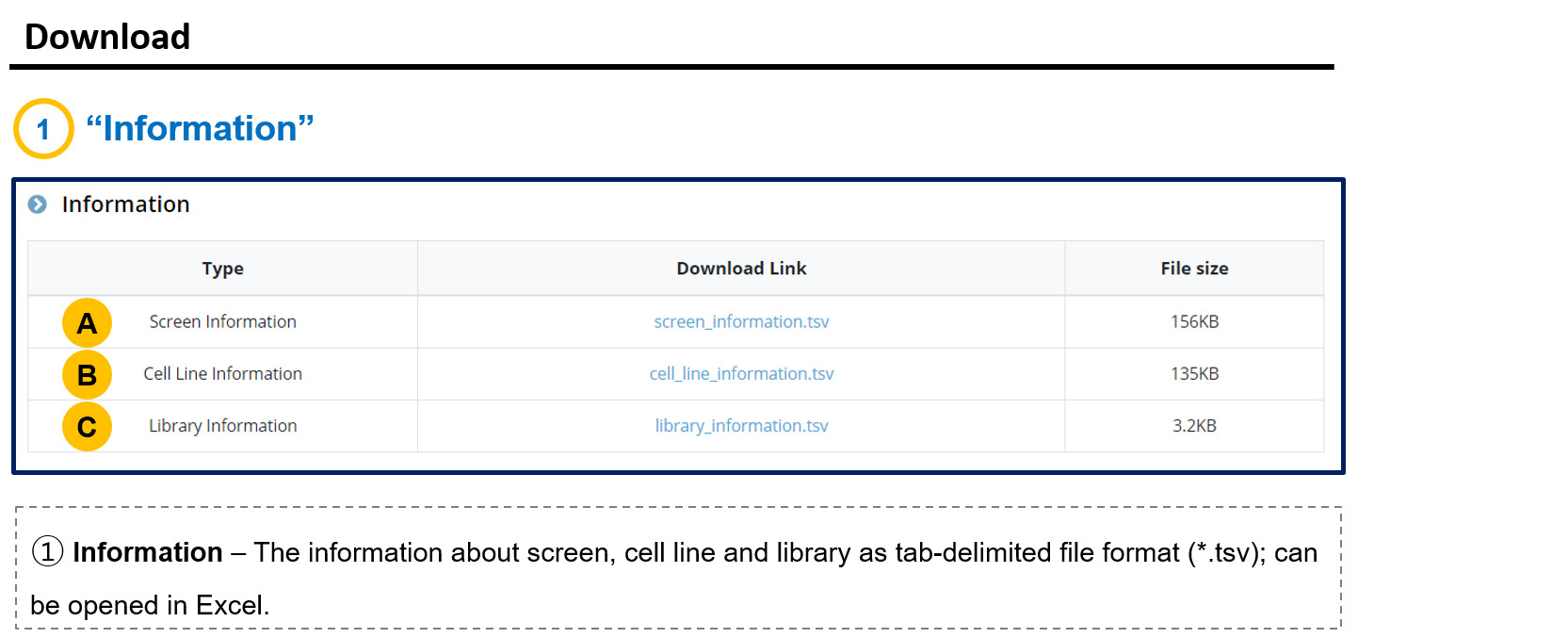
- 3. Download
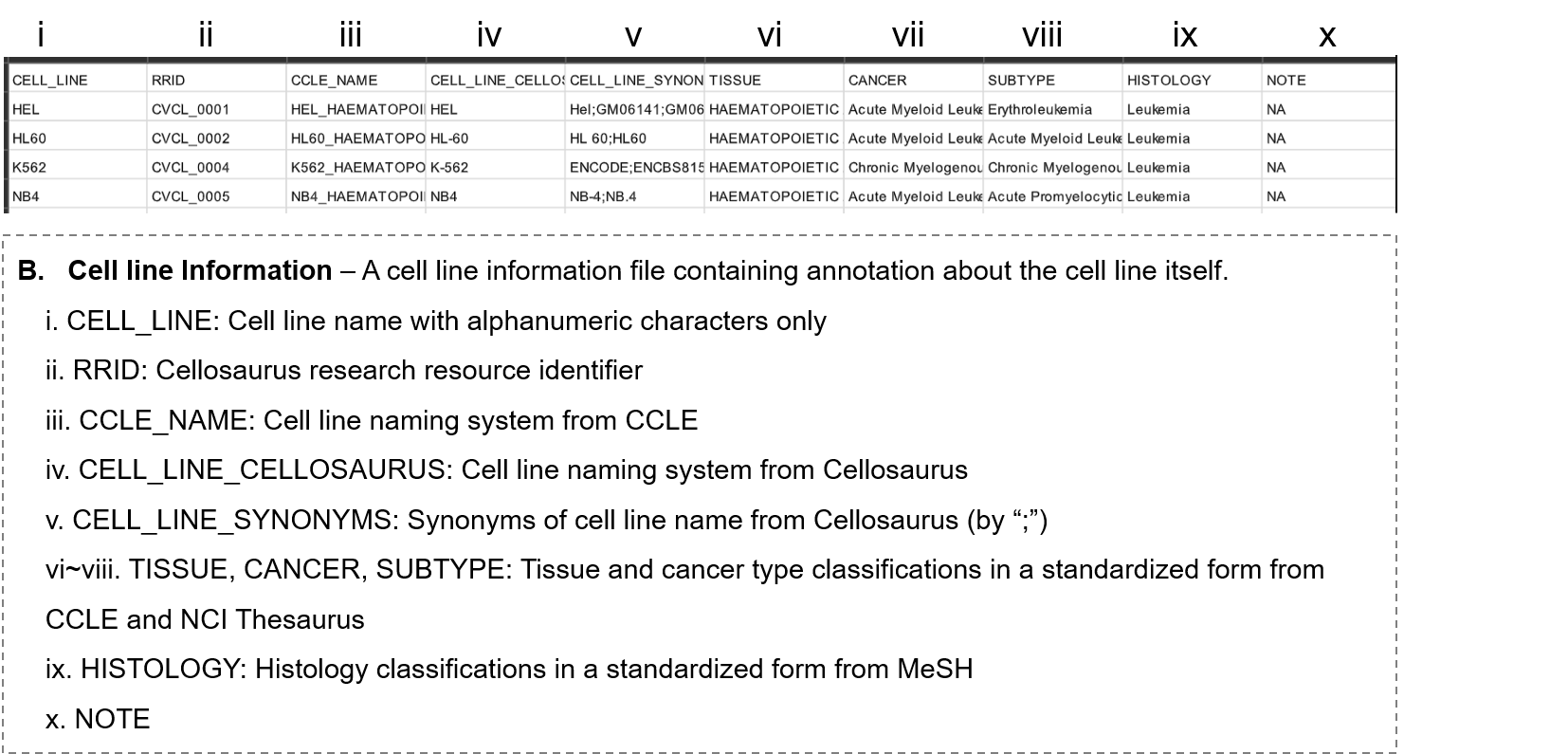
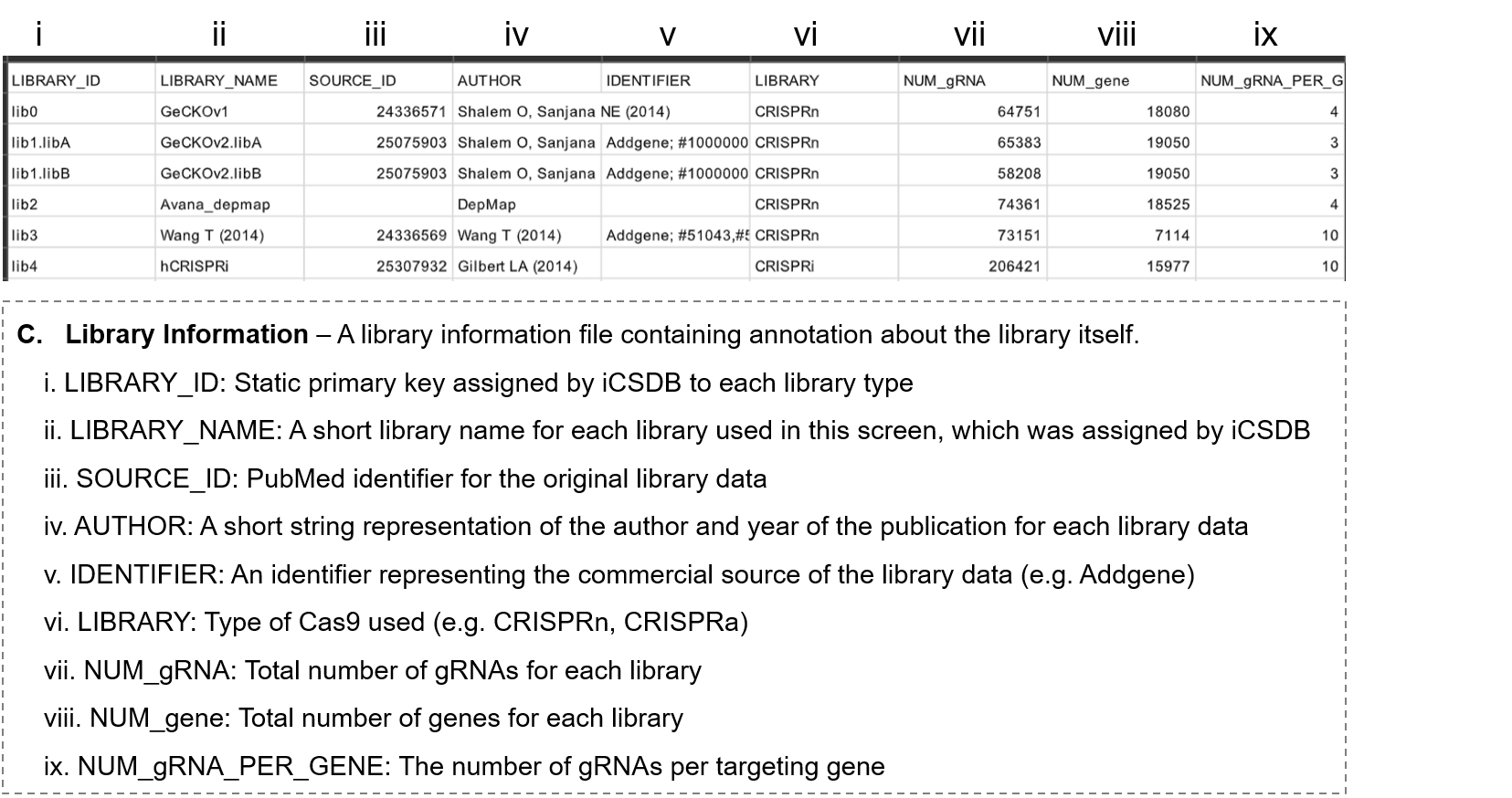
- 3-1. Information
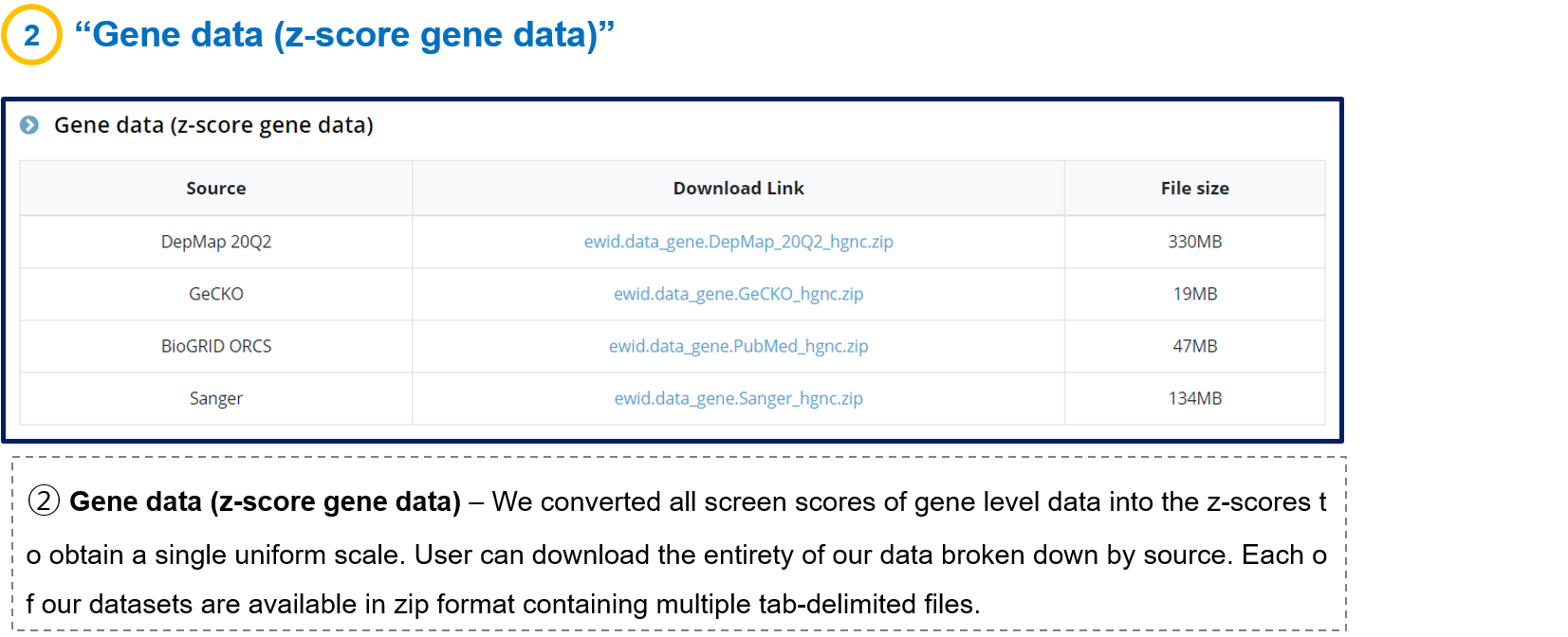
- 3-2. Gene data (z-score gene data)
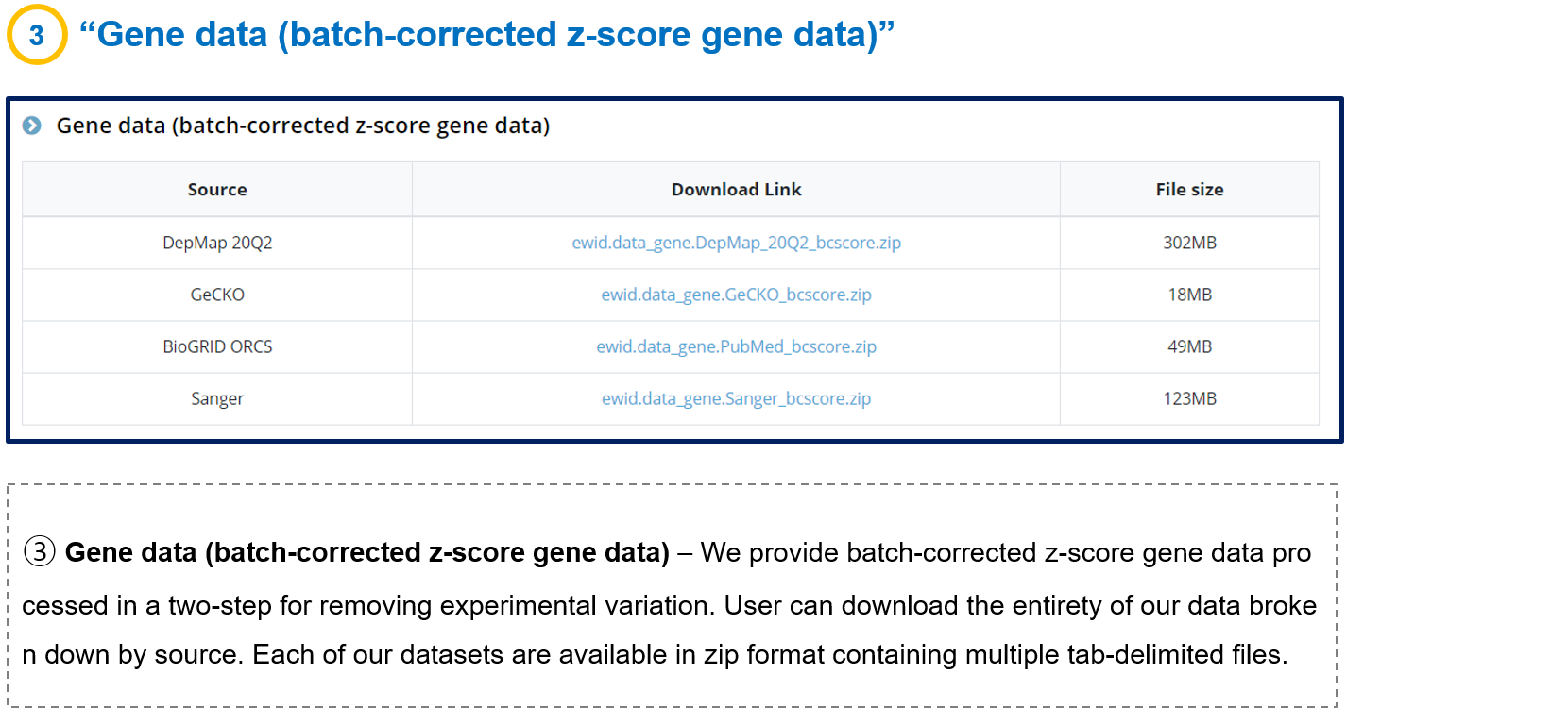
- 3-3. Gene data (batch-corrected z-score gene data)
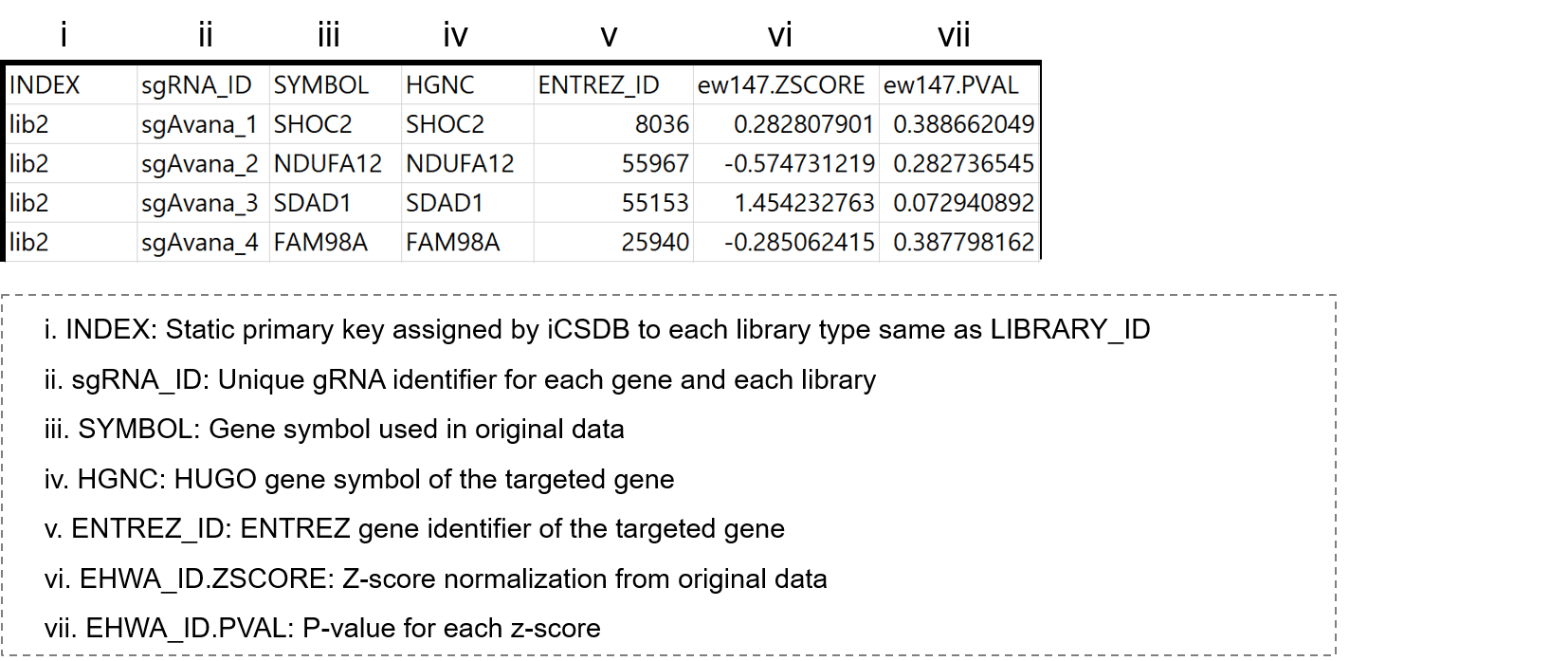
- 3-4. gRNA data
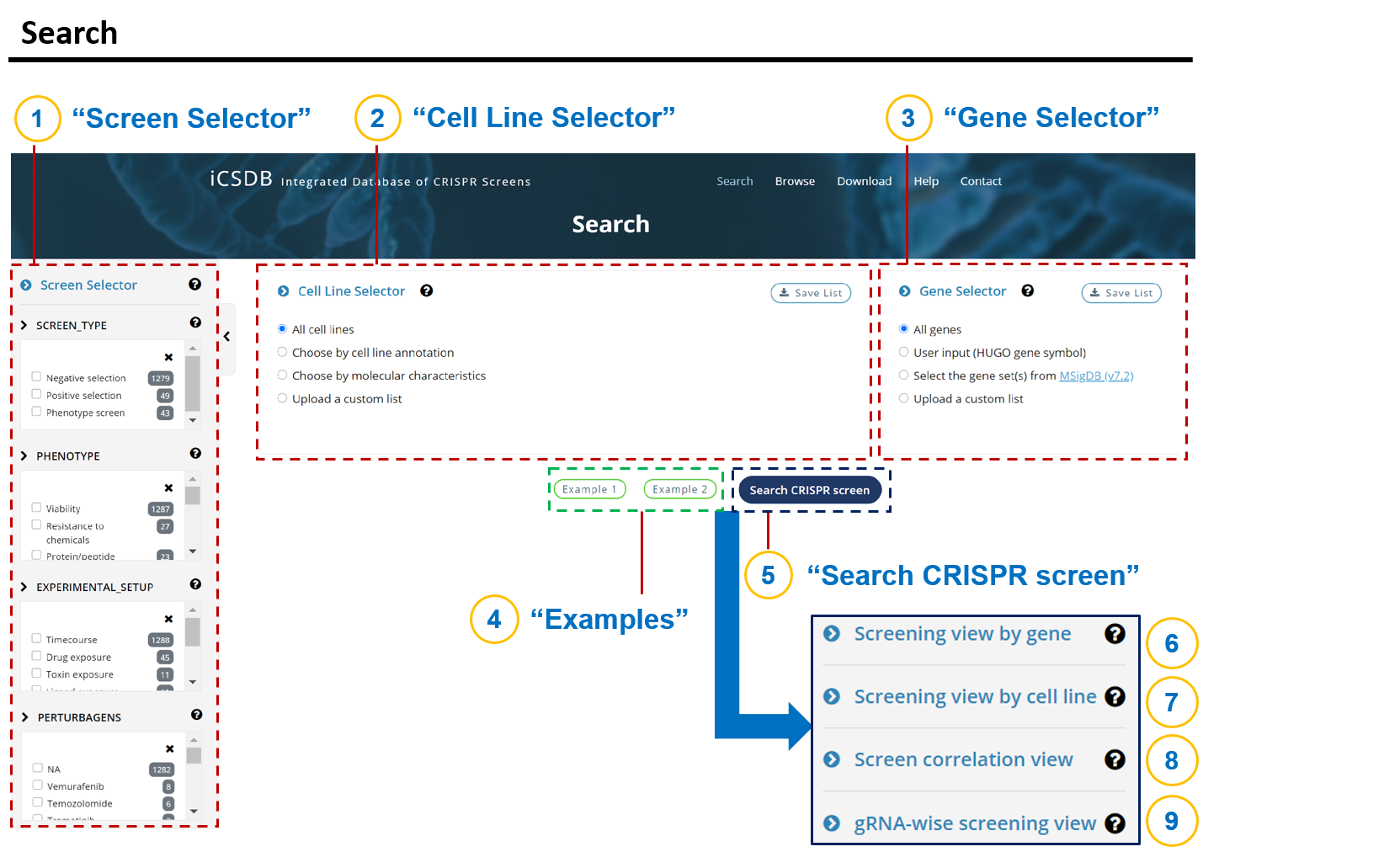











The database search needs three inputs as designed in the Search page.
1) Screen
2) Cell line
3) Gene
The default selections are all screens, all cell lines, and all genes.
Users may narrow down any search category using the selectors.
Selecting all screens, all cell lines, and all genes is fast because the result is pre-computed and pre-loaded.
Other selections or queries are dealt with in real-time and they may yield a huge amount of output, whose transfer to the user's computer may take some time.
This is especially the case when the number of cell lines (and genes) is several hundred (and thousands).
Please be patient. The website is not down.
In this case, we want to find genes that are sensitive to CRISPR knockout (i.e. negative selection) in the lung adenocarcinoma cell lines.
1) Select the screen type as "negative selection".
2) Select the cell lines for lung adenocarcinoma using cell line classifications. Lung adenocarcinoma can be found in the cancer subtype category.
3) Press the "Search CRISPR screen" button.
In the output page, the screening view by gene figure shows the essentiality score in each cell line (batch-corrected z-score) in ascending order.
Choosing genes on the left panel will add the genes in the picture.
The detailed score can be examined by downloading the excel file.
You can use the "Choose by molecular characteristics" feature in the Cell Line Selector.
1) Choose MutationType
2) Search "KRAS" in the Gene Name, which will give cell lines with KRAS mutations
3) You may further choose a specific mutation (e.g. G12D) in the "AA mutation".
4) Press the "Save List" button.
Similarly, other molecular characteristics such as expression, copy number, and gene fusions may be used to select the cell lines of interest.
We provide the MSigDB database to help guide the gene selection.
(H) Hallmark gene sets or (C5) Gene Ontology gene sets are recommended, but any choice is fine.
Screening views by gene and by gRNA-wise are available.
Choose the genes and cell lines of interest in the graphical interface, and download the CSV file to see the details.
Note that the number of genes and cell lines in the gRNA-wise view is limited by 5 due to the speed and space.