About ChimerDB
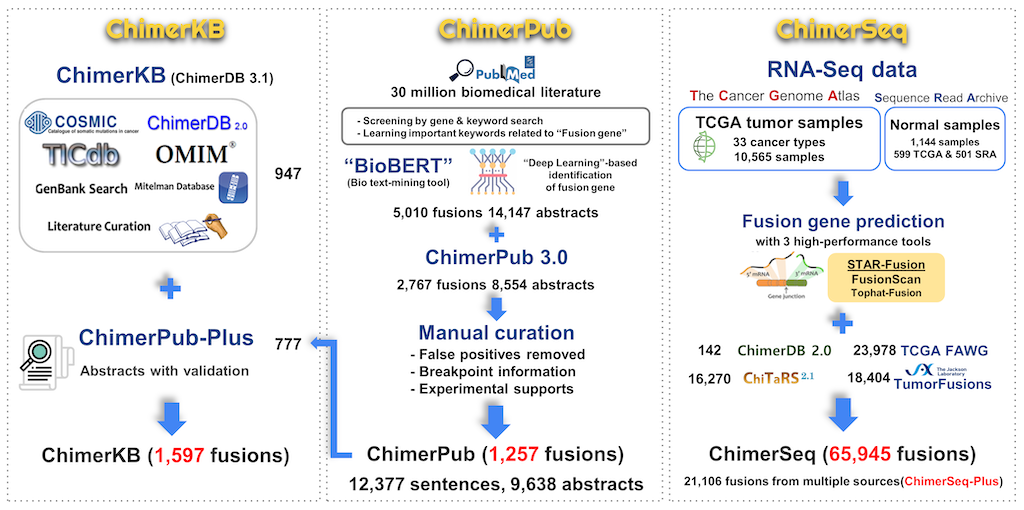
ChimerDB is a comprehensive database of fusion genes encompassing analysis of deep sequencing data (ChimerSeq) and text mining of PubMed publications (ChimerPub) with extensive manual annotations (ChimerKB). This update version 4.0 contains 67,610 fusion genes. Major improvements are as follows:
- ChimerSeq covers all TCGA data and provides the ChimerSeq-Plus subset as highly reliable fusions.
- Quality of ChimerPub content was greatly enhanced by applying a new "deep learning"-based text-mining method followed by extensive manual curation.
- ChimerPub-Plus that contains fusions with literature and experimental supports increased ChimerKB’s content by ~50%.
- ChimerSeq module supports diverse visualization tools including fusion structure view, gene expression plot, STRING network view, and circos plot.
Citing ChimerDB
Please cite "ChimerDB 4.0: an updated and expanded database of fusion genes" submitted
Previous versions
ChimerDB 3.0: an enhanced database of fusion genes with cancer transcriptome and literature data mining. Lee M, Lee K, Yu N, Jang I, Kang J, Lee S. Nucleic Acids Res. 2017 Jan 4;45(Database issue):D784-9.Web site
ChimerDB 2.0: a knowledgebase for fusion genes updated. Kim P, Yoon S, Kim N, Lee S, Ko M, Lee H, Kang H, Kim J, Lee S. Nucleic Acids Res. 2010 Jan;38(Database issue):D81-5.
ChimerDB: a knowledgebase for fusion sequences. Kim N, Kim P, Nam S, Shin S, Lee S. Nucleic Acids Res. 2006 Jan 1;34(Database issue):D21-4.